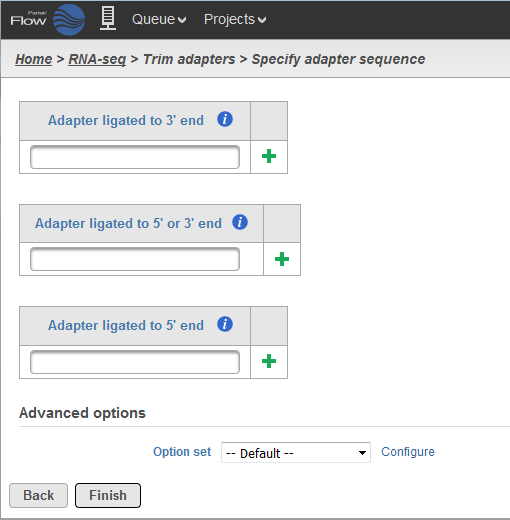
GitHub - naturalis/galaxy-tool-cutadapt-sequence-trimmer: Use cutadapt to trim and discard reads and read sections.
Cutadapt remoYes adapter sequences from high-throughput sequencing reads Introduction Implementation Features
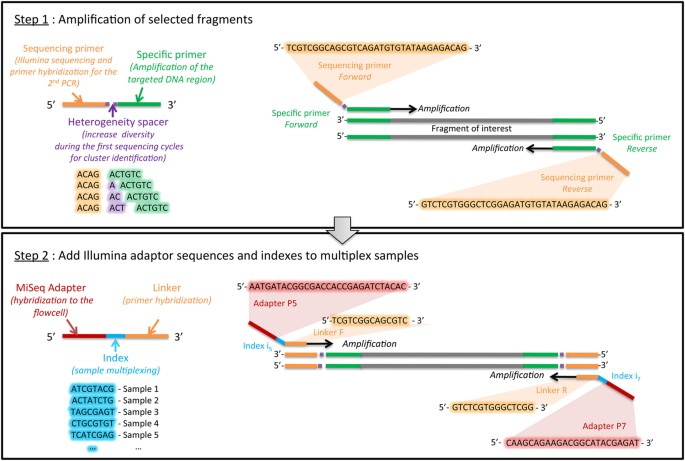
High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy | Scientific Reports
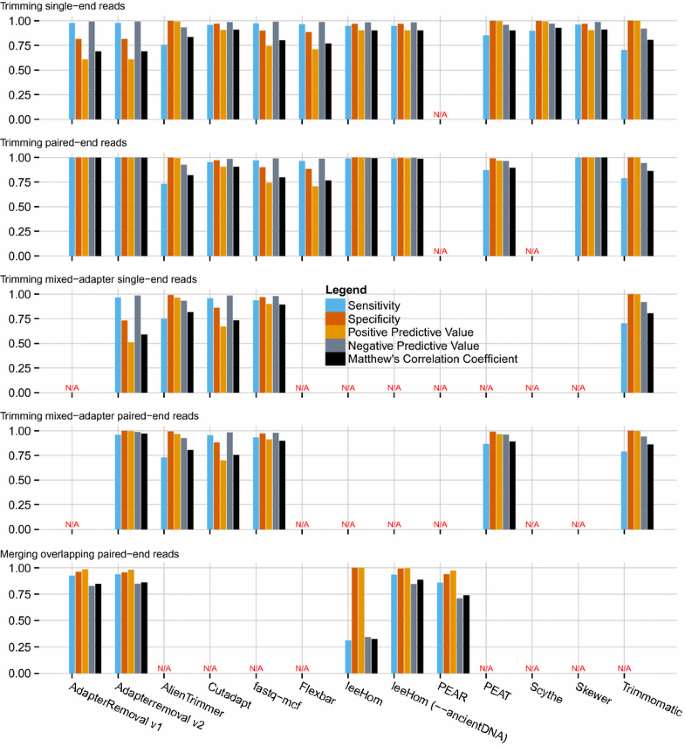
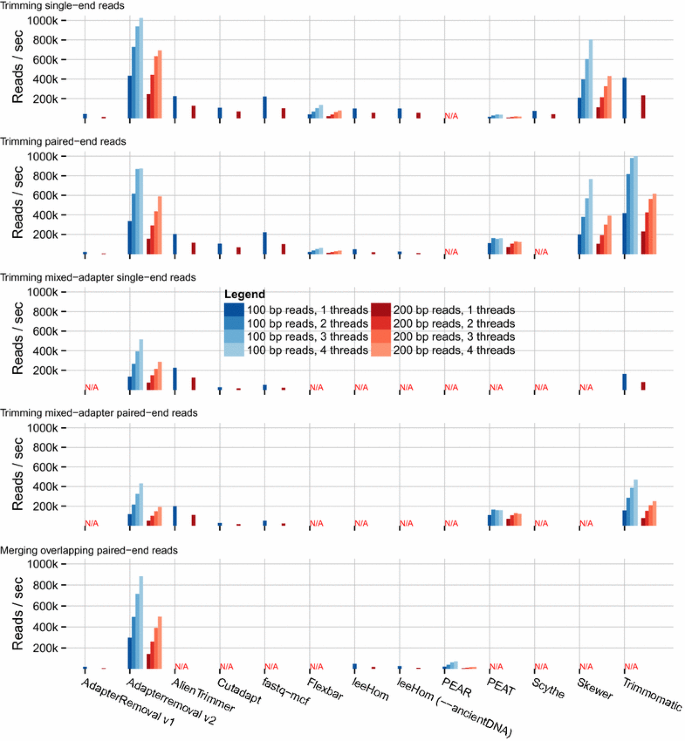
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text
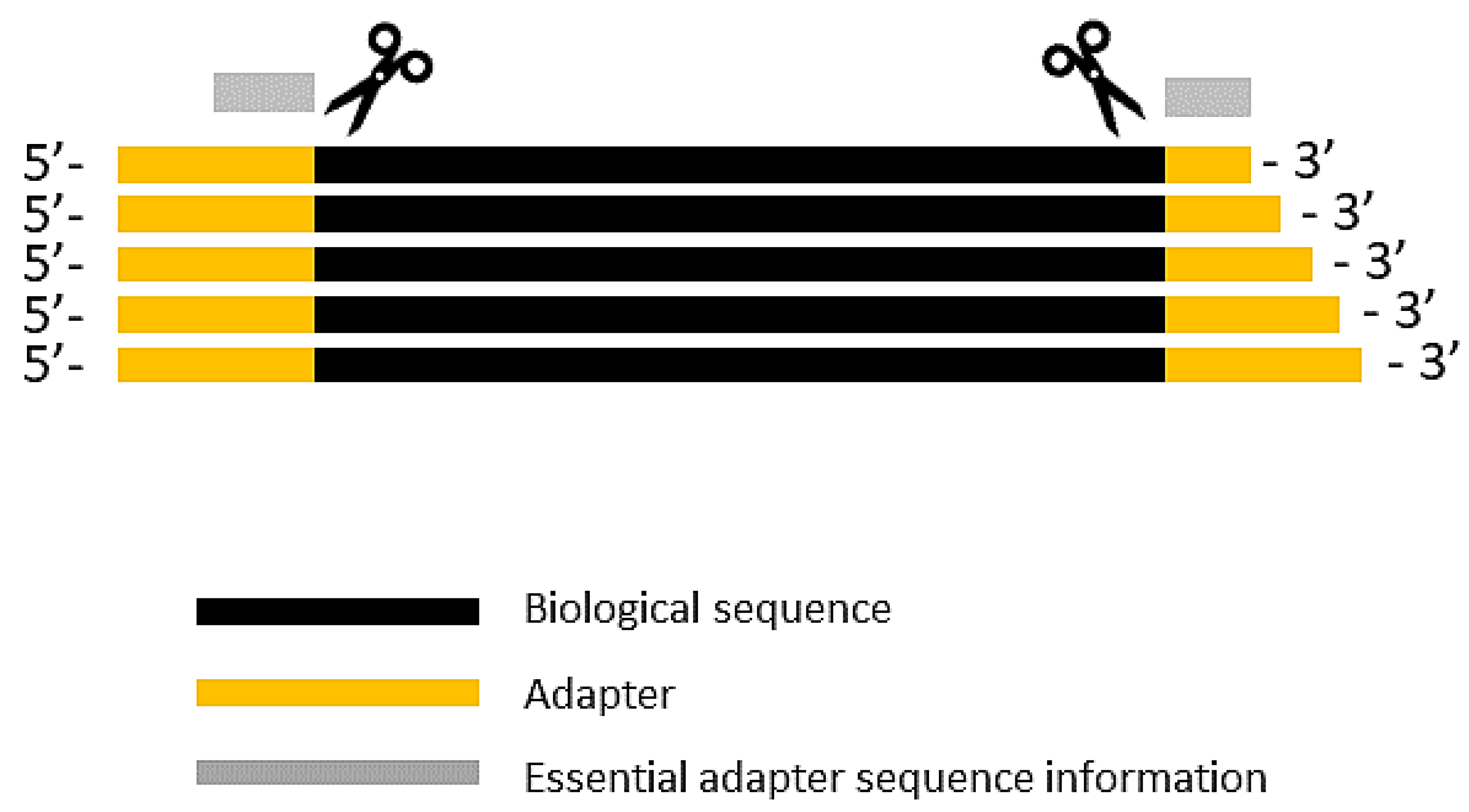
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

Kraken: A set of tools for quality control and analysis of high-throughput sequence data☆ | Semantic Scholar
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text









![2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ] 2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/14041/1/fig-2-full.png)